Cat. #155083
pcPPT-mPGK-attR-sLPmCherry-WPRE vector
Cat. #: 155083
Sub-type: pRRL Lentiviral backbone
Availability: 3-5 days
Bacterial Resistance: Ampicillin
Selectable Markers: mCherry expression in mammalian cells
This fee is applicable only for non-profit organisations. If you are a for-profit organisation or a researcher working on commercially-sponsored academic research, you will need to contact our licensing team for a commercial use license.
Contributor
Inventor: Luigi Ombrato ; Ilaria Malanchi
Institute: The Francis Crick Institute
Tool Details
*FOR RESEARCH USE ONLY
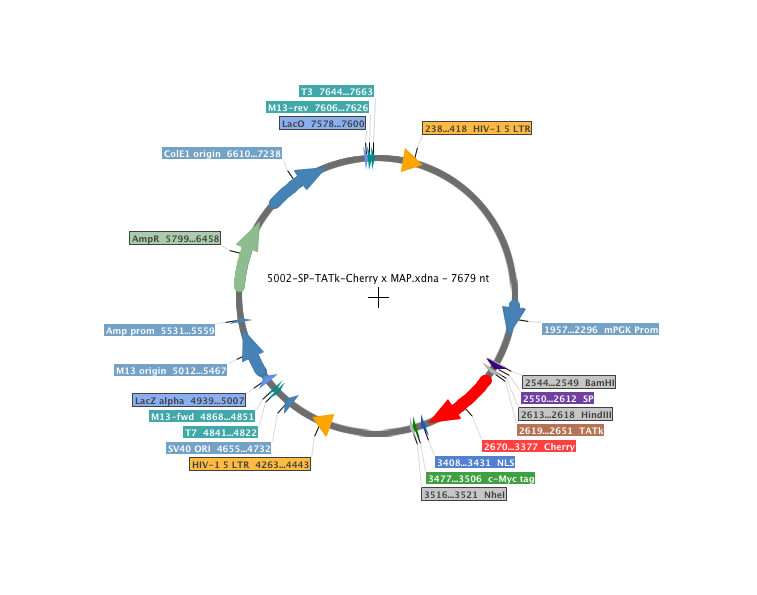
- Tool name: pcPPT-mPGK-attR-sLPmCherry-WPRE vector
- Alternate name: Metastatic-niche labelling system, psLPmCherry
- Tool sub type: pRRL Lentiviral backbone
- Backbone size without insert: 6800
- Bacterial resistance: Ampicillin
- Selectable markers: mCherry expression in mammalian cells
- Description: A metastatic-niche labelling system. This lentiviral backbone plasmid contains a soluble peptide (SP) and a modified TAT peptide cloned upstream of the mCherry gene, under the control of a mouse PGK promoter (sLPmCherry). Cell lines stably transfected with the plasmid express and release a cell-penetrating fluorescent protein, which is taken up by neighbouring cells and enables spatial identification of the local metastatic cellular environment. Using this system, tissue cells with low representation in the metastatic niche can be identified and characterised within the bulk tissue. To highlight its potential, Ombrato et al., Nature 2019, applied this strategy to study the cellular environment of metastatic breast cancer cells in the lung. They engineered 4T1 breast cancer cells to co-express the sLPmCherry and GFP; these cells are referred to as labelling-4T1 cells. In vitro, sLPmCherry protein secreted by labelling-4T1 cells re-enters the cells, as indicated by changes in the intracellular localisation of the red fluorescence. sLPmCherry protein is also taken up by unlabelled cells, both in co-culture with labelling-4T1 cells and when cultured in medium conditioned by labelling-4T1 cells (LCM). Upon uptake into a cell, sLPmCherry fluorescence has an intracellular half-life of 43 hours and is localised in CD63+ multi-lamellar bodies (lysosomal-like structures) where, owing to its high photostability, it retains high fluorescence intensity. Fractionation of LCM shows that only the soluble fraction retains labelling activity, whereas the extracellular vesicles, a proportion of which contain sLPmCherry, do not show labelling activity in vitro. In vivo, intravenous injection of labelling-4T1 cells (GFP+mCherry+) into syngeneic BALB/c mice to induce lung metastases efficiently labels surrounding host tissue cells (GFP-mCherry+), penetrating approximately five cell layers. This enables specific discrimination of host cells in close proximity to cancer cells from distal lung cells (GFP-mCherry-) using fluorescence-activated cell sorting (FACS). Notably, when micro-metastases grew larger, the number of mCherry+-niche cells in the tissue remained proportional to the number of metastatic cells.
Handling
- Storage conditions: -80° C
- Shipping conditions: Dry Ice
Application Details
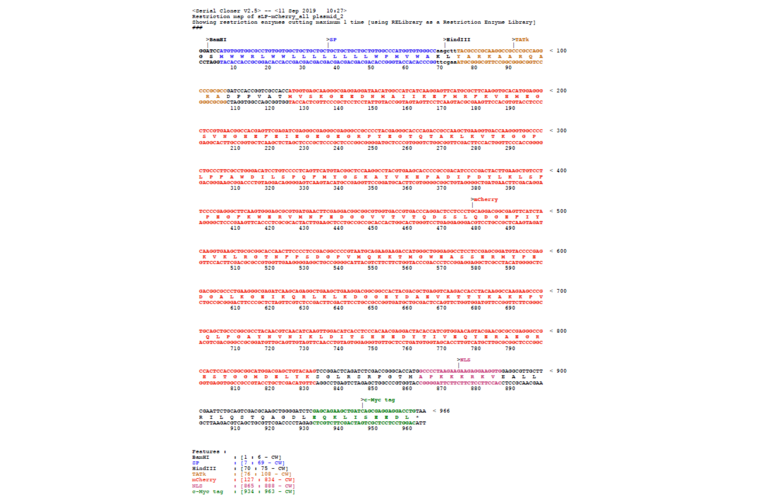
- Application notes: Protocol: Generation of in vivo labelling tumour cell lines by Luigi Ombrato and Ilaria MalanchiThe sLP-Cherry can be subcloned by using BamHI and NheI restriction sites.Expression of the sLP-mCherry fusion is controlled by the mouse PGK promoter. The originators of the plasmid have introduced the plasmid into a human breast cancer cell line, MDA-MB-231, and sLP-mCherry was expressed.
References
- Generation of in vivo labelling tumour cell lines by Luigi Ombrato and Ilaria Malanchi. Protocol Exchange, DOI: 10.21203/rs.2.14720/v1
- Ombrato et al. 2019. Nature. PMID: 31462798.