At CancerTools, we’re proud to power global cancer research through one of the most comprehensive collections of cancer cell lines. Our portfolio includes 1,500 well-characterised models developed by leading cancer scientists and institutions, including pioneering research funded by Cancer Research UK. Each cell line carries with it a story of scientific innovation and impact. As a non-profit organisation that is part of Cancer Research UK, every purchase supports our shared mission to beat cancer. In this post, we highlight some of our most requested and influential cell lines – trusted by academic and industry researchers alike to drive discovery and innovation.
MB49: the original bladder cancer model
When it comes to bladder cancer research, MB49 remains one of the most referenced and relied-upon cell lines. CancerTools is the exclusive, verified source of the authentic MB49 line, obtained directly from originator Dr Leonard Franks at Cancer Research UK’s Lincoln’s Inn Fields (1).
MB49’s value lies in its ability to capture tumour-immune dynamics in an immunocompetent, syngeneic system. Because it recapitulates key features of the human bladder cancer microenvironment, you can use it to explore immune evasion, tumour progression, and therapeutic response with physiological relevance. Additionally, its demonstrated sensitivity to checkpoint inhibitors, including anti-PD1, makes it especially powerful for immunotherapy research and development (2). MB49-luciferase – a modified derivative of MB49 – also continues to be a vital model for immunotherapy research in immunocompetent C57BL/6 mice. It enables real-time in vivo tracking of tumour progression and metastasis, offering researchers crucial insights into cancer biology and immunology.
For researchers seeking a reliable, traceable and translationally meaningful bladder cancer model, MB49 offers a dependable foundation.
Explore the original MB49 line.
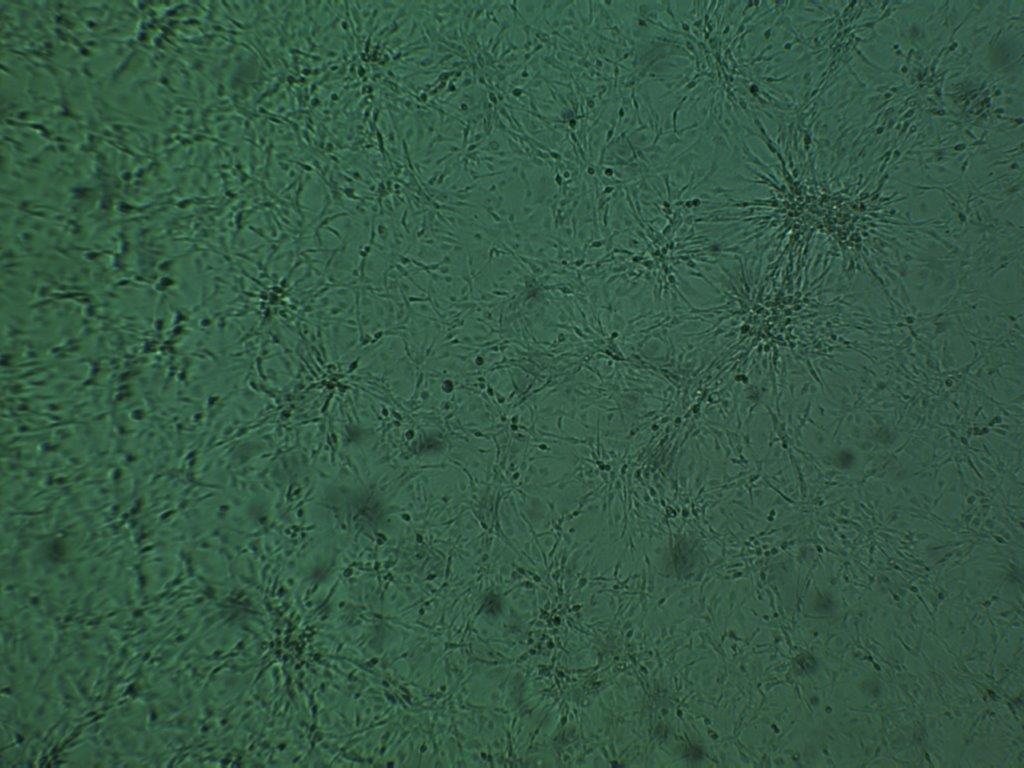
MB49 cell line, passage 3.
UM-UC panel: capturing bladder cancer diversity
Developed by Prof. H. Barton Grossman and Dr Anita Sabichi, the UM-UC panel comprises 11 patient-derived urothelial carcinoma cell lines (3). This panel is widely used for modelling bladder cancer heterogeneity and supports research ranging from drug resistance to biomarker discovery.
What makes the UM-UC panel so special? Each cell line – all well-characterised in literature – originates from a distinct patient tumour, capturing a spectrum of genetic and clinical backgrounds. This diversity and its close reflection of patient tumours allow researchers to explore everything from tumour biology and drug resistance to biomarker discovery (4). Because the panel includes well-documented tumourigenicity data, it supports both in vitro assays and in vivo modelling – a major advantage for those building on translational pipelines.
If you’re exploring tumour biology, screening therapeutics, or developing biomarkers, this panel offers a robust platform for confident experimental design.
Access the UM-UC bladder cancer panel.
PEO series: tracing drug resistance in ovarian cancer
Developed by Prof. Simon Langdon at Cancer Research UK’s Edinburgh Centre, the PEO series stands out as a powerful model for studying the evolution of drug resistance in high-grade serous ovarian cancer (HGSC) (5). Derived from a single patient at multiple treatment stages, these lines allow you to explore how ovarian cancer adapts across relapse and therapeutic pressure.
PEO1 arises from a relapse after prior platinum exposure and carries a BRCA2 Y1655X mutation that removes full-length BRCA2, resulting in homologous recombination (HR) deficiency (6). This makes it a robust, and widely used model for exploring platinum sensitivity, PARP inhibitor response, and the vulnerabilities associated with defective DNA repair. PEO4, taken at a later relapse, carries a BRCA2 reversion mutation that restores HR function – a defining feature associated with resistance to both platinum and PARP inhibitors6. PEO6, sampled later in the patient’s disease course shows further molecular changes, including restored BRCA2 function and treatment resistance – making it especially useful for modelling late-relapse HGSC (7).
To extend this series, in vitro–selected derivatives such as PEO1-OR (olaparib-resistant) and PEO1-CDDP (cisplatin-resistant) provide complementary models for studying how platinum and PARP inhibitor resistance emerges under therapeutic pressure. Together, these lines form a powerful toolkit for researchers investigating treatment resistance, synthetic lethality, and DNA repair dynamics in ovarian cancer (8).
Advance your ovarian cancer research with the PEO series.
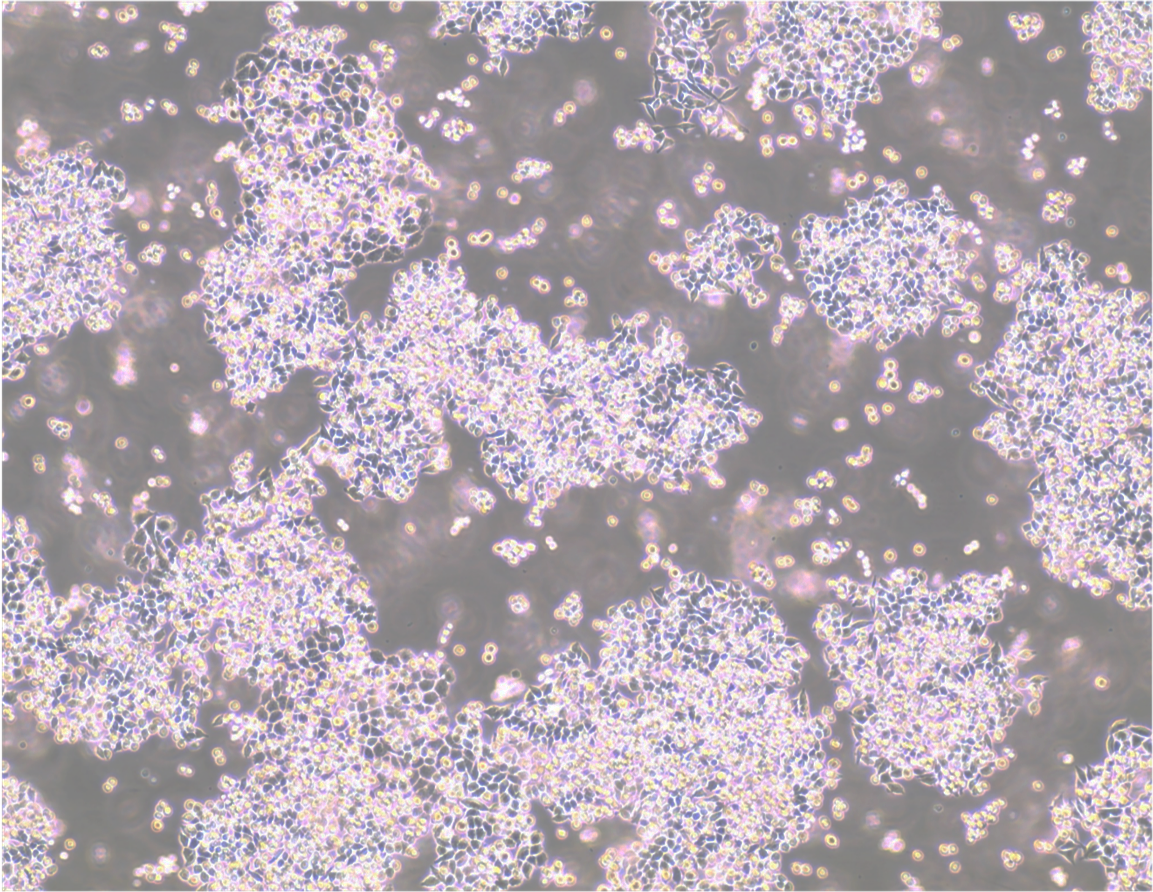
PEO1 cell line. 3 days post plating. Image courtesy of the European Collection of Authenticated cell Cultures (ECACC).
A2780: a foundational model for ovarian cancer
The A2780 cell line, originally developed by Dr Stuart Aaronson at the National Cancer Institute, remains one of the most widely used models for ovarian cancer research (9). Derived from an untreated patient with endometrioid adenocarcinoma, A2780 is well known for its intrinsic sensitivity to cisplatin, providing a dependable baseline for studies exploring cancer genetics and therapeutic toxicity.
Importantly, A2780 serves as the parent line for the widely used A2780cis (cisplatin-resistant) and A2780ADR (adriamycin-resistant) derivatives (10). This makes it a cornerstone model for researchers investigating how resistance emerges, how it can be overcome, and how new agents perform across sensitive-resistant pairs. Its ability to grow in both monolayer and suspension, coupled with reliable tumour formation in immunodeficient mice, allows seamless integration between in vitro assays and in vivo validation.
For researchers benchmarking therapies, validating mechanisms, or comparing resistance states, A2780 offers a reproducible and translationally relevant system.
Explore the A2780 lineage.
CMT series: in vivo tumourigenesis models for lung cancer metastasis
The CMT series – CMT 64/ CMT 64/61, CMT 167, and CMT 170 – originating from work by Prof. Peter Riddle at Cancer Research UK’s Lincoln’s Inn Fields, provides a dependable suite of murine alveolar lung cancer models widely used to study tumour growth, metastasis, and immune interactions (11). Their consistent morphology and stable metastases make them trusted tools for researchers seeking reproducible results.
CMT 64 is particularly valued as a robust in vivo tumourigenesis model, offering stable growth in culture and in lung metastases after subcutaneous inoculation. This reliability allows researchers to assess drug candidates, investigate tumour progression, and generate high-quality efficacy data on immunoresistance and metastasis (12,13). Building on this model, CMT 167 was developed with enhanced metastatic potential, making it especially powerful for modelling aggressive disease, mapping immune evasion, and evaluating therapeutic response in translational relevant contexts (14-16).
For scientists aiming to model clinically meaningful aspects of lung cancer progression and accelerate preclinical development, the CMT series delivers a proven and impactful set of tools.
Advance your lung cancer studies with the CMT series.
MCF7 and T47D: models for endocrine-resistant breast cancer research
Developed by Dr Anne Lykkesfeldt at the Danish Cancer Society, the anti-oestrogen-resistant MCF7 and T47D derivatives are derived from the foundational oestrogen-receptor-positive (ER+) breast cancer cell lines, MCF7 and T47D, providing clinically relevant models for studying how tumours adapt to hormone therapy. These variants demonstrate resistance to hormone-dependent breast cancer treatments, making them powerful tools for investigating resistance mechanisms and for supporting the development of novel predictive biomarkers for therapy response (17-21).
Anti-oestrogen–resistant derivatives, including tamoxifen-resistant lines such as MCF7/TAMR-1, provide a clinically relevant system for investigating how tumours evade hormone therapies. These lines allow researchers to uncover molecular drivers of resistance, test next-generation endocrine therapies, and generate reproducible, translationally meaningful results that support the development of more effective therapeutic strategies.
Additionally, gene-edited MCF7 derivative lines with site-specific BRCA1 promoter hypermethylation enable the study of BRCA1-silenced tumourigenesis without relying on genetic mutations. Together, this suite of breast cancer models offers a robust platform for advancing research into endocrine resistance and BRCA-related mechanisms with confidence.
Access the MCF7 and T47D models.

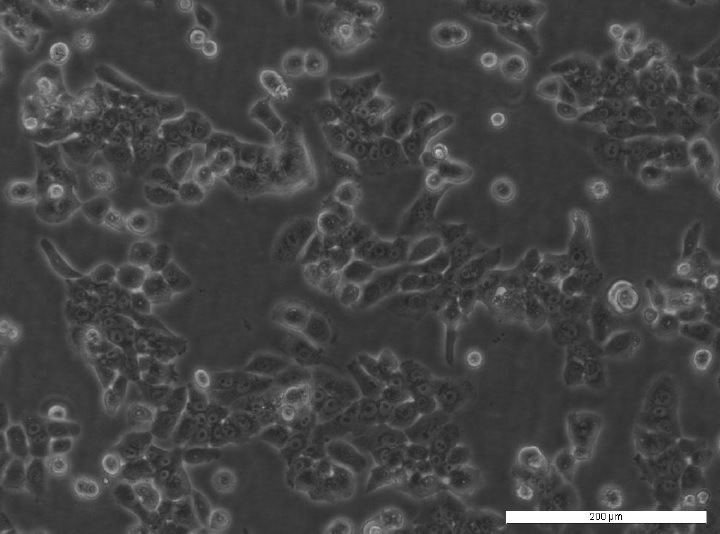
MC7F/TAMR-8 cell line. Image courtesy of the European Collection of Authenticated cell Cultures (ECACC).
HCT 116 BRCA2-/-: colorectal cancer models for translational research
The HCT 116 BRCA2-/- clones, developed by Dr. Carlos Caldas at Cancer Research UK Cambridge Institute, are derived from the well-characterised human colorectal carcinoma line HCT 116. These homozygous knockout lines, including Clone 46 and Clone 42, feature targeted disruption of BRCA2, resulting in the loss of Rad51 foci, chromosomal rearrangements, and heightened sensitivity to DNA-damaging agents and PARP1 inhibitors (22).
By offering a BRCA2-deficient background that can be experimentally compared against wild-type HCT 116, these models enable researchers to generate reproducible, clinically relevant preclinical data. They are widely used to support investigations into DNA repair mechanisms, synthetic lethality, and therapeutic vulnerabilities, offering insights that are directly translatable to drug development and precision oncology.
For teams studying BRCA2-driven biology or testing new therapeutic strategies, the HCT 116 BRCA2-/- clones provide a robust platform for generating translationally meaningful results, supporting confident decision-making in preclinical pipelines.
Explore the HCT116 BRCA2-/- clones.
Why researchers choose CancerTools
At CancerTools, we provide peer-reviewed, expert-developed research products that deliver reproducible, high-quality, and meaningful results. By sourcing or depositing tools through us, scientists contribute to a global ecosystem that accelerates cancer discovery, ensuring their work has lasting impact and drives the next generation of cancer breakthroughs.
We make access to these research tools seamless: streamlined licensing, reliable distribution, and worldwide availability remove barriers so researchers can focus on science, not logistics. As a not-for-profit, every accessed tool, including cell lines, supports both the originating inventor or institute and Cancer Research UK, reinvesting in future discoveries while creating a legacy that shapes the field.
Access the tools trusted by leading cancer researchers today. Explore our cell line catalogue and find the models that will advance your next breakthrough.
References
- Summerhayes IC et al. 1979. Journal of the National Cancer Institute. 62(4):1017–1023. PMID: 107359
- Vandeveer AJ et al. 2016. Cancer Immunology Research. 4(5):452–462. PMID: 26921031
- Sabichi A et al. 2006. The Journal of Urology. 175(3):1133–1137. PMID: 16469639
- Zuiverloon TCM et al. 2018. Bladder Cancer. 4(2):169–183. PMID: 29732388
- Langdon SP et al. 1988. Cancer Research. 48(21):6166–6172. PMID: 3167863
- Sakai W et al. 2009. Cancer Research. 69(16):6381–6386. PMID: 19654294
- Biegala L et al. 2023. Cells. 12(7):1038. PMID: 37048111
- Greenwood W et al. 2019. Clinical Cancer Research. 25(8):2471–2482. PMID: 30651275
- Parker RJ et al. 1991. Journal of Clinical Investigation. 87(3):772–777. PMID: 1999494
- Dutil J et al. 2019. Cancer Research. 79(7):1263–1273. PMID: 30894373
- Franks LM et al. 1976. Cancer Research. 36(3):1049–1055. PMID: 1253168
- Rincón E et al. 2017. Oncotarget. 8(28):45415–45431. PMID: 28525366
- Miyashita N et al. 2021. Sci Rep. 17;11(1):22380. PMID: 34789779
- Evans et al. 2009. Cancer Res. 69(5):1733-8. PMID: 19208832
- Li et al. 2017. Cancer Immunol Res. 9: 767-777. PMID: 28819064
- Bullock et al. 2019. Life Sci Alliance. 27;2(3): e201900328. PMID: 31133614
- Lykkesfeldt AE et al. 1986. British Journal of Cancer. 53(1):29–35. PMID: 3947513
- Kirkegaard T et al. 2014. Cancer Letters. 344(1):90–100. PMID: 24513268
- Thrane et al. 2015. Oncogene. 34:4199–4210. PMID: 25362855
- Elias et al. 2015. Oncogene. 34:1919–1927. PMID: 24882577
- Larsen et al. 2015. PLoS One. 23;10(2):e0118346. PMID: 25706943
- Xu H et al. 2014. Journal of Pathology. 234(3):386–397. PMID: 25043256