Introduction
Post-translational modifications (PTMs) impact eukaryotic cell diversity by altering protein activity and coordinating cell signalling networks. PTMs are the chemical changes that occur after DNA has been transcribed by RNA into a protein. Defects within PTMs, can cause numerous diseases including:
- cancers where histone methylation causes the repression of tumour suppressor genes;
- autoimmune disorders such as rheumatoid arthritis, caused by increased histone methylation; and
- Alzheimer’s disease, where histone acetylation and deacetylation play a role in memory formation.
By increasing our understanding of the mechanisms of PTMs, we can develop new treatments for these diseases.
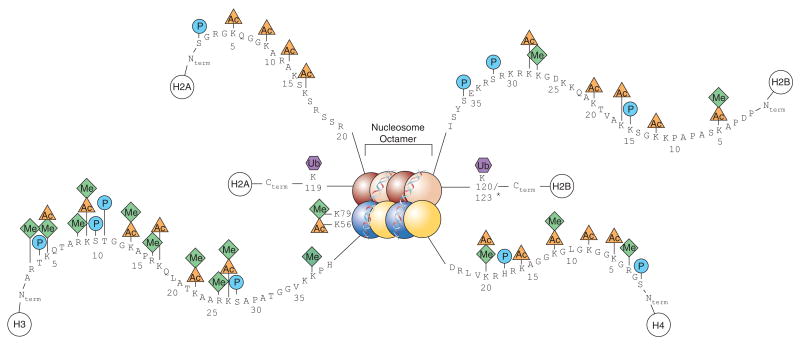
Image: A diagram showing a full nucleosome with including the histone PTMs on their respective histones. Taken from Keppler, Brian & Archer, Trevor. (2008). Chromatin-modifying enzymes as therapeutic targets – Part 1. Expert opinion on therapeutic targets. 12. 1301-12. 10.1517/14728222.12.10.1301.
PTMs play a crucial role in histone modifications, as they regulate the ability of histones to condense DNA from single strands to tightly wrapped super coiled DNA within nucleosomes. These nucleosomes consist of two dimers of histone H2A-H2B and a tetramer of H3-H4 histones (see Fig.1) and all contain a simple helix turn helix motif and a long amino acid “tail”. PTMs to the long amino acid “tail” change the histone’s function, leading to histone involvement in transcription, DNA repair and replication. These modifications also form the basis of the five different histone PTM families. This article will focus on these five histone PTM families and their associated impacts:
Methylation
Histone methylation is a common PTM that causes differing numbers of methyl groups to attach to a single histone amino acid. Errors in the location and number of methyl groups bonded to this amino acid lead to developmental defects, increased cancer progression and alterations in the DNA repair pathway. Recently Zheng et al., have shown that two methyltransferases (MLL3 and MLL4) associated with mono-methylation of H3K4, have distinct mechanisms, with MLL3 found in gastrulation and MLL4 in lung development. Based on this research, specific inhibitors or activators targeted to each transferase can be designed, providing unique tools to use in precision medicine targeting blood-cell and developmental disorders.
Gene expression is continually suppressed and increased through the activity of MLL methyltransferases, and demethylases. One well characterised demethylase, KDM4, is now a promising target in cancer therapeutics. Disfunction of the KDM4 enzyme leads to sustained proliferation and genomic instability, which in breast cancer can be overexpressed by approximately 60%. This over-expression means KDM4 inhibitors are an interesting therapeutic target with new chemicals being studied regularly for their ability to rebalance gene expression.
Acetylation
Knowledge of histone acetylation and deacetylation has become important in understanding transcriptional regulation. In recent years, inhibitors of histone deacetylases (HDAC), which cause genetic repression through the removal of acetyl groups, have become popular in cancer drug development. For example, in patients with stage 4 neuroblastoma, over-expression of HDAC10 leads to cytotoxic drug resistance resulting in poor survivability of a patient.
Research has shown that the inhibition of this protein increases cell sensitisation to these cytotoxic drugs, which could result in higher numbers of successful cancer treatments. Common essential genes found within this drug resistance pathway, are also found within apoptosis. These genes need to be tightly regulated using acetylation and deacetylation, otherwise uncontrollable cell proliferation occurs. In December 2020, Rajan et al., published a review into Histone Acetylation-/Methylation-Mediated Apoptotic Gene Regulation in Hepatocellular Carcinoma, showing that an increase in acetylation on histone H2B, H4 and hypoacetylation of histone H4 leads to a progression in cellular apoptosis. This can also be seen when the loss of H4K16 deacetylati
Knowledge of histone acetylation and deacetylation has become important in understanding transcriptional regulation. In recent years, inhibitors of histone deacetylases (HDAC), which cause genetic repression through the removal of acetyl groups, have become popular in cancer drug development. For example, in patients with stage 4 neuroblastoma, over-expression of HDAC10 leads to cytotoxic drug resistance resulting in poor survivability of a patient.
Research has shown that the inhibition of this protein increases cell sensitisation to these cytotoxic drugs, which could result in higher numbers of successful cancer treatments. Common essential genes found within this drug resistance pathway, are also found within apoptosis. These genes need to be tightly regulated using acetylation and deacetylation, otherwise uncontrollable cell proliferation occurs. In December 2020, Rajan et al., published a review into Histone Acetylation-/Methylation-Mediated Apoptotic Gene Regulation in Hepatocellular Carcinoma, showing that an increase in acetylation on histone H2B, H4 and hypoacetylation of histone H4 leads to a progression in cellular apoptosis. This can also be seen when the loss of H4K16 deacetylation specifically causes inhibition and deviation from the normal cell death pathway in Non-alcoholic Steatohepatitis. By increasing our understanding of HDACs and their inhibitors, advancements in cancer treatments can be achieved that build on the already successful therapeutic toolbox.
on specifically causes inhibition and deviation from the normal cell death pathway in Non–alcoholic Steatohepatitis. By increasing our understanding of HDACs and their inhibitors, advancements in cancer treatments can be achieved that build on the already successful therapeutic toolbox.
Ubiquitylation and Sumoylation
It is estimated that 5–15% of H2A and 1–2% of H2B are conjugated with ubiquitin in vertebrate cells. Ubiquitination plays a role in the regulation of transcription and histone ubiquitylation can define a histone’s activity and protein-protein interactions. In April 2021, cross talk between ubiquitylation, acetylation and methylation was identified. DOT1, a methyltransferase with involvement in leukaemia, had its activity increased by H4K16 acetylation and H2B ubiquitylation. This fundamental research provides further mechanistic detail into leukaemia development that may result in cancer therapeutics.
Histone SUMOylation occurs on all four core histones, the linker histone H1 and histone variants H2A.Z and H2A.X. Although it was discovered over a decade ago, the direct effects of histone SUMOylation are only recently being explored. While histone SUMOylation was originally designated as a marker of transcription repression by competing with other PTMs on histones, it has now been established that SUMOylation is a fine tuner of transcription and DNA damage where it can both act as a repressor or activator. Discussions on druggable sumoylation factories that provide opportunities for clinical interventions within cancers, neurodegeneration and viral infections are now taking place.
Phosphorylation
Histone phosphorylation plays a major role in both transcription and DNA repair. A common measurement of DNA damage is the amount of histone H2A.XS139phospho (γH2AX) present within a cell. γH2AX notifies the cell of a double stranded break and results in downstream signalling of repair proteins and cell cycle stalling. This use of γH2AX as a measuring tool of DNA damage has allowed studies into glomerular diseases where type VI collagen can be deposited as large nodular lesions within the body.
As well as being used as a signal of DNA damage, the actual repair mechanism and signalling pathways linked to γH2AX are still being researched. A recent Nature Communications publication by Dobersch et al., shows that the presence of H2A.X phosphorylation at sites of DNA damage, removes the methylation groups off DNA, allowing for transcriptional activity at the gene of interest. This adds further mechanistic detail to an already complex process of which comes first, PTMs on a histone or the recruitment of proteins associated with sensing DNA damage. By answering this question, researchers can improve the cells ability to fix damaged cells before they become oncogenic.
Conclusion
Whilst PTMs on histones have been heavily characterised, their roles have only become more complex, with some both activating and repressing transcription. With 8 histones making a nucleosome and 30 million nucleosomes per cell, the cross talk between all these PTMs is complex. This article shares only a glimpse into the latest research, highlighting a few of the new applications and potential uses of histone PTMs. The increased focus on histone PTMs and the subsequent discoveries could help provide new avenues into potential treatments for various cancers, and other associated diseases in the near future.