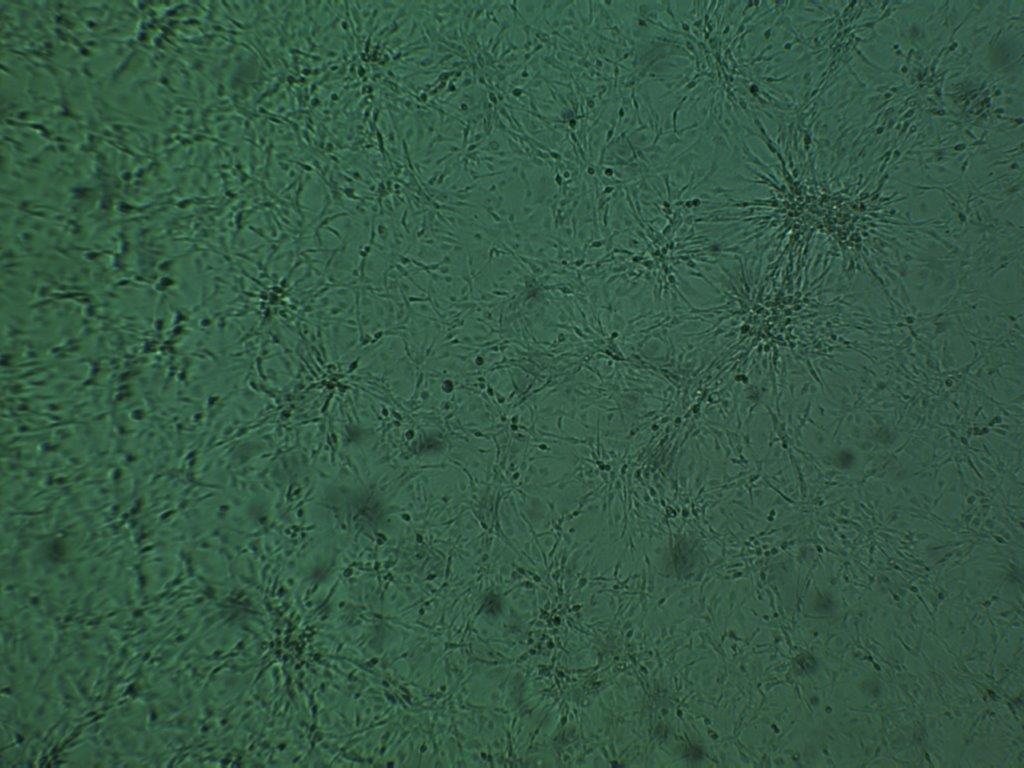
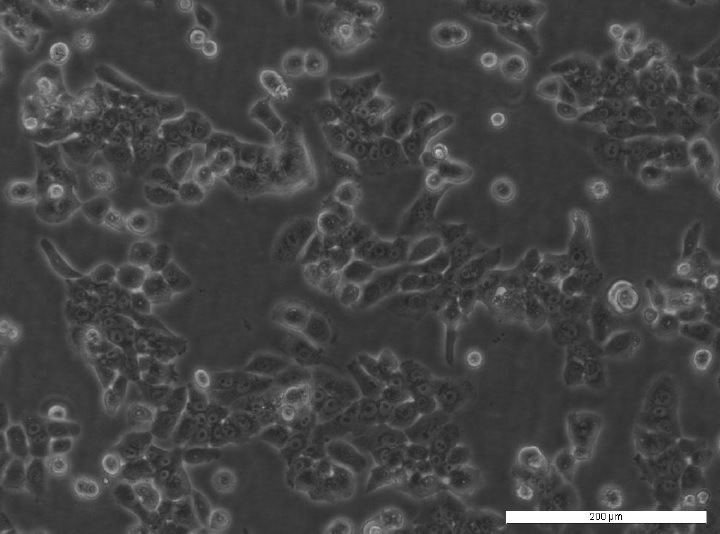
The research tool: GATA3-eGFP reporter cell line
GATA3, a zinc finger transcription factor, is associated with numerous types of cancer in which its level of expression is critical. Drugs that modulate GATA3 expression are of particular interest, which is why a high-throughput screening tool is an important addition to the research tool portfolio. Matthew Holley, Emeritus Professor at the University of Sheffield, has shared insights about such one-of-a-kind tool – the GATA3-eGFP reporter cell line, and other in vitro models he has developed throughout his research- which he has generously contributed to CancerTools.org.
”There are no other cell lines with this potential for the study of GATA3 in cancer research.
Emeritus Prof. Holley

The contributor
University of Sheffield; School of Biosciences
GATA3 in development and cancer
GATA3 is one of the GATA family transcriptional factors; zinc finger proteins that bind the consensus DNA sequence (T/A)GATA(A/G) of a target gene promoter. It is highly conserved among vertebrates and expressed in various tissues and cell lineages, such as immune cells, adrenal glands, placenta, kidneys, skin and breast tissue, inner ear, hair follicles, and nervous system.
GATA3 is responsible for regulating transcription during both development and cell differentiation, and its expression level is crucial at embryonic and postnatal stages. The importance is apparent from examples like early embryonic lethality due to GATA3 deletion, HDR syndrome associated with GATA3 haploinsufficiency, and the fact that Gata3 has been implicated in tumorigenesis. Previous studies have shown that it is involved in T-cell neoplasms, antagonizes cancer progression in PTEN-deficient prostates, and represents a useful marker for luminal category tumours in breast cancer.
GATA3-eGFP reporter cell line for drug discovery
In 2009, Prof. Holley’s group published results from a gene array analysis they conducted using inner ear cell lines [1]. They showed a clear, previously unknown, functional link between GATA3 and the Akt signalling pathway; increased activity of which is often associated with tumour progression and therapies resistance. The pathway is essential for regulating cell survival and proliferation, and aberrations in its activation are linked to several human cancers, including breast, lung, ovarian, and prostate cancers.
Prof. Holley delved into exploring how to modulate gata3 expression, recognising its fundamental role in significant pathways associated with cancer progression as well as in the development of various organs and the nervous system.

Image: Inventing institution: University of Sheffield, UK
The GATA3-eGFP cell line was established by the cross of two well-characterised transgenic mouse lines. One mouse line stably expressed the H-2Kb-tsA58 transgene in which a temperature-sensitive variant of the SV40 large T-antigen was expressed under the control of an inducible promoter driven by gamma-interferon. The second, GATA3eGFP BAC-transgenic mouse line expressed eGFP under the control of the GATA3 promoter. The clones were selected according to the characteristics of the original tissue.
The GATA3-eGFP reporter cell line expresses GATA3-eGFP via an enhancer that is expressed in a pattern very closely resembling that of native GATA3 during mouse embryonic development. Fusion of GATA3 enhancer to eGFP is a key feature of this cell line allowing high-throughput screening of drugs that modulate the expression level of gata3.
First high content screening tests with established GATA3 modulators have shown effectiveness at the anticipated concentrations in the GATA3-eGFP cell line. The tool stands out as the only GATA3 reporter line capable of efficiently screening extrinsic factors influencing GATA3 expression in high-throughput systems.
The key objective of this case study is to show the high potential of this tool for those research laboratories interested in understanding GATA3’s role in various processes in cancer and other diseases.
Other in vitro models
For many years, Prof. Holley dedicated his research efforts to understand development and functioning of the inner ear in mammals, with an emphasis on exploring potential therapies for hearing loss.
Mammalian hair cells located in the inner ear are sensory cells that detect sound, gravity, and acceleration. Progressive loss of the hair cells is one of the main causes of deafness. Hearing loss is widespread and mostly irreversible as the mammalian cochlea is unable to regenerate hair cells that are lost, unlike amphibians and birds.
Mammalian auditory research is complicated by the fact that the very small numbers of auditory sensory epithelial cells do not proliferate postnatally or in vitro, which makes them experimentally inaccessible. Thus, Prof. Holley and his colleagues developed a number of in vitro models for the differentiation of sensory hair cells and sensory nerves.
The established inner ear cell lines are derived from known cell populations during specific times of mouse inner ear development. They have been shown to correlate with gene expression profiles of the location from which they were derived. The lines are also conditionally immortal and can be grown under proliferating or differentiating conditions.
These cell lines were generously contributed to CancerTools.org and used for exploring the possibility of inner ear regeneration through both cell transplantation and the activation of early developmental processes [2, 3].
”I believe in open access to all reagents where possible. They are costly to make and their application can advance research as well as return some investment back to the charitable funding agencies that enabled their production.
Emeritus Prof. Holley

Image: Expression of the β4 integrin subunit in VOT-N33 and VOT-E36. (Adopted from Lawoko-Kerali et. al. 2004. Dev Dyn. 231(4):801-14. PMID: 15499550 Fig.2 B, C)
Conclusion:
At CancerTools.org we continue to work with Cancer scientists who can deposit research tools they have developed in their labs, including cell lines, antibodies, organoids, mouse models, cell culture media, small molecules and other state-of-the-art technologies, into our biorepository etc.